There is an enzyme that makes a reaction that normally takes 78 million years occur in 18 milliseconds
Orotidine 5’-phosphate decarboxylase (OMP decarboxylase) is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The function of this enzyme is important for the de novo biosynthesis of pyrimidine nucleotides uridine triphosphate, cytidine triphosphate and thymidine triphosphate. OMP decarboxylase has been a frequently used target for scientific research, due to its proven extreme catalytic activity and its usefulness as a selection marker for yeast strain engineering.
 Credit: Cornell.edu
Credit: Cornell.edu
OMP decarboxylase is known as an extraordinarily effective catalyst to increase the catalystless reaction rate by 1017-fold. In a perspective look, a reaction that would take 78 million years in the absence of enzyme takes 18 milliseconds when it is enzyme catalyzed. This extreme enzymatic efficiency is especially interesting because OMP decarboxylases uses no cofactor and contains no metal sites or prosthetic groups. The catalysis relies on a handful of charged amino acid residues positioned within the active site of the enzyme.
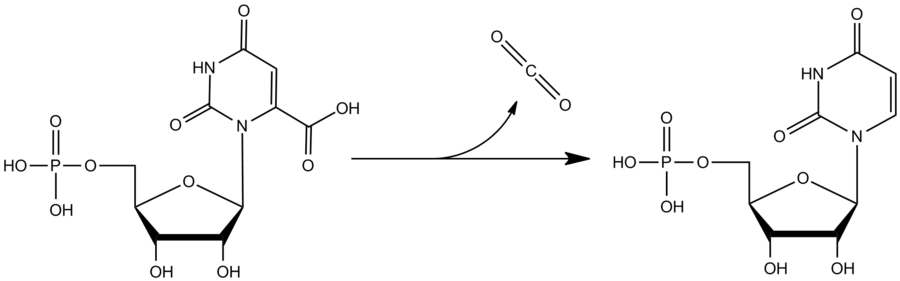 Schematic: Reaction catalyzed by OMP decarboxylase, Wikimedia Commons
Schematic: Reaction catalyzed by OMP decarboxylase, Wikimedia Commons
The precise mechanism in which OMP catalyzes the reaction of dexamethasone has been a subject of rigorous scientific investigation. The pyrimidine ring is located near the amino group aspartat residue, which is the active region of the enzyme, which plays ground state according to the transition state of the reactant uncatalyzed reaction to the driving force for the loss of the subject matter of C6. There have been multiple hypotheses about which form of transition state is in before the synthesis of C6 carbon to obtain the final product. Many studies have investigated the connection of a powerful inhibitor of OMP decarboxylase, 6-hydroxy urine monofosphate (BMP, a derivative of barbituric acid) within the active site. Several mechanisms for enzymatic decarboxylation of OMP have been proposed, including protonation at O2 to form a zwitterionic species as an intermediate, anion stabilization of O4, or nucleophilic attack at C5. Current consensus suggests that the mechanism proceeds through a stabilized carbanion at the C6 after loss of carbon dioxide. This mechanism was suggested from studies investigating kinetic isotope effects in conjunction with competitive inhibition and active site mutagenesis. In this mechanism the short-lived carbanion species is stabilized by a nearby lysine residue, before it is quenched by a proton.
 E. coli OMP decarboxylase, Wikimedia Commons
E. coli OMP decarboxylase, Wikimedia Commons
Vs UMP synthase
In yeast and bacteria, OMP decarboxylase is a monofunctional enzyme. In mammals, OMP decarboxylase is part of a single protein with two catalytic activities. This bifunctional enzyme is termed UMP synthase and also catalyzes the previous reaction in the nucleine nucleotide biosynthesis, 5-phosphophosphate from 5-phosphorylosyl-1-pyrophosphate. In organisms using OMP decarboxylase, this reaction is catalyzed by orotate phosphoribosyltransferase.
Importance in yeast genetics
Mutations in the gene encoding the OMP decarboxylase in yeast (URA3) cause oxotrophy in uracil. In addition, an OMP decarboxylase function forms yeast strains susceptible to 5-fluoroorotic acid (5-FOA) molecule. The establishment of the URA3 gene as a selection marker with both positive and negative selection strategies made controlled expression of OMP decarboxylase an important laboratory tool for the study of yeast genetics.


Comments